BDB-Lab Winter 2021 Update
GMGCv1 paper is out and much more
Note: We will be holding online open office hours on January 13 (noon UTC). Registration is required to minimize spam, but is otherwise open for all in this link.
The Global Microbial Gene Catalog v1.0 paper is out!
This is our Winter 2021 quarterly update. We will send four of these a year, (roughly) coinciding with the two equinoxes and the two solstices. Every four months, we highlight one of the projects in the group and give short updates on everything else that has been happening.
This time, we release it a bit later to avoid the holidays around the Winter solstice, but this is still the Winter 2021 edition.
Winter 2021 Focus: Global Microbial Gene Catalog v1
This time the focus is on the Global Microbial Gene Catalog (GMGC)
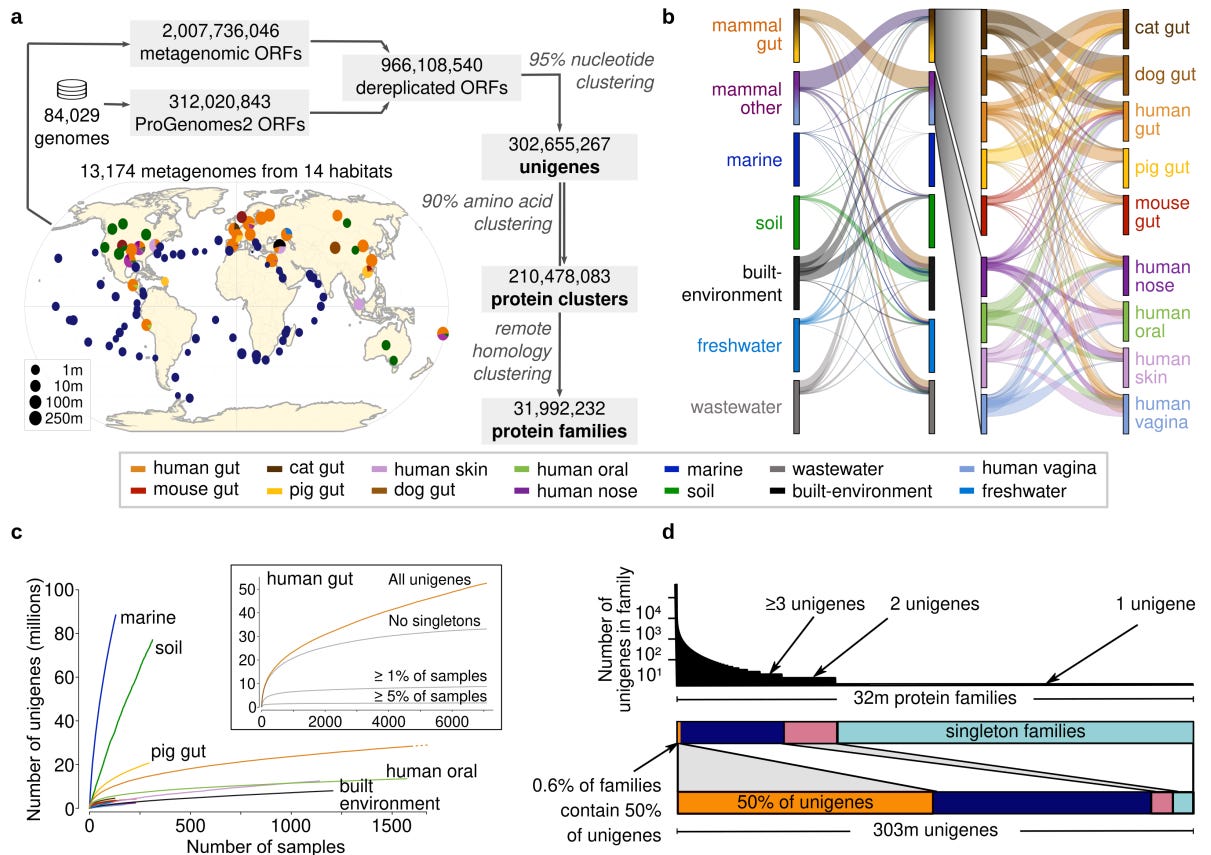
The structure of prokaryotic genes. In this work, we collated >10,000 metagenomes and almost 100,000 isolate genomes to produce a gene catalog containing >300 million unigenes (species-level clusters, at 95% nucleotide identity). These were further clustered into 32 million protein families (broadly-defined, any statistically significant, detectable, homology).
Analyses revealed that (1) most genes are habitat-specific—which is true for both unigenes and protein families—and that (2) most of this diversity appears to be the product of neutral (or nearly-neutral) evolution rather than adaptation to local environmental conditions.
The global microbiome is a single system. Even though one of our results is that most genes are habitat-specific, this is not true in all cases and it depends on the resolution you look at: protein families are more cosmopolitan than unigenes; and species are more cosmopolitan than strains (even between very similar habitats1).
Therefore, this paper (and our work) aims to study the global microbiome as a single system. The field has traditionally been split into studies of different habitats, so that some scientists study the human microbiome, others study the ocean, so that each habitat has its own specialists. This approach has been very fruitful, but ignores that everything is connected.
GMGC (Global Microbial Gene Catalog) as a resource. The GMGC is also a resource for others to use. There is an interactive website (https://gmgc.embl.de/), which also includes some novel sequence querying algorithms to enable fast (interactive) searching over 300 million gene sequences. Advanced users can download all the data at https://git.embl.de/coelho/GMGC10.data. We are also making the catalog (and habitat specific subcatalogs) available for use with NGLess.
The future of GMGC. The fact that we refer to GMGC, version 1 is a hint that we consider this just the first version in a long-term project. We are already building GMGCv2. We are also using the resource to investigate the distribution of sequences and organisms across the global microbiome. For example, this is the basis of our participation in the EMBARK Project.
Other BDB-Lab Updates
People. Yan Liang recently joined BDB-Lab as a graduate researcher working on our global microbiome analyses. To know him better, visit our page of members.
Svetlana and Anna finally made it to Shanghai!
Manuscripts & abstracts. GMGC.v1 is published in Nature.
The “Open code pledge” was submitted to metarXiv: https://osf.io/preprints/metaarxiv/vrwm7/
Hui’s paper describing the EXPERT tool for microbial source tracking was updated on bioRxiv.
Conferences. Shaojun presented the SemiBin at MVIF.2.
The ResFinderFG.v2 was presented at #ICCMg6 and #RICAI2021 including Svetlana & Luis (EMBARK collaboration).
Tutorial. The tutorial for SemiBin was presented on November 2nd and we released a video demonstration of Jug in our new youtube channel. These online hands-on sessions train people on using our tools, while also providing valuable feedback that we can use to enhance usability.
We intend to announce more tutorials in the near future. Sign up for the tutorials mailing-list to keep informed.
Online resources & tools. Macrel tool has a new released version (v.1.1), in which we eliminated R dependencies, fixed the classifier list for PyPI, added support for compressed files other than gzip (.xz and .bz2), included more extensive testing and fixed a few bugs. After all these improvements, Macrel now is faster than before (~3.5x). You can find Macrel on bioconda.
AMPSphere has its first online test version. The resource website is now available as a beta and feedback is highly appreciated. There you will be able to access different antimicrobial peptide sequences, their genes and locations as well as their microbial sources and environmental distribution. Sequence search tools are also available.
A new version of SemiBin (v0.5) was released in GitHub with several improvements. Reclustering is now the default, which should return slightly better bins. GTDB lazy downloading is now performed even when using a non-standard directory, while the standard cache directory implements the CACHEDIR.TAG protocol. Several internal improvements made SemiBin more efficient, with a lower memory usage, especially when using a pretrained model.
Website & blog. We have a revamped website: https://www.big-data-biology.org/
A new blogpost is now up on the BDB-Lab blog. In this post, Anna Vines discusses her research conducted at BDB-Lab during her internship, finding evidence of cryptic antimicrobial peptides production by prokaryotes in the environment.
Microbiome Forum. Hui and Anna joined the steering committee of the Microbiome Virtual International Forum. MVIF is a recurring bite-sized alternative to a multi-day microbiome conference. Each event is replicated shortly thereafter to reach a global audience and build a global microbiome network. MVIF is free: everyone is welcome to discuss microbiome science!
Webinar. At the December EMBARK2021 seminar, Svetlana talked about how we explore the global resistome using the global microbial gene catalog.
Other. Eldin Jašarević presented his research to our group.
Looking Forward
We are now looking for remote internships again! If you are interested in our remote internship program (or any other topic that we work on), we will be holding online open office hours on January 13 (noon UTC). Registration is required to minimize spam, but is otherwise open for all in this link.
We had earlier already shown how dogs and humans share species, but strains appear to be host-specific, which in a completely different way, explains some of the observations we report in the SemiBin preprint.


